Fixed particles¶
Overview¶
Questions¶
How can I create arbitrarily shaped barriers?
Objectives¶
Show how to place particles to form arbitrary barriers.
Demonstrate the technique for HPMC and MD simulations.
Boilerplate code¶
[1]:
import gsd.hoomd
import hoomd
import numpy
The render function in the next (hidden) cell will render the system state using fresnel. Find the source in the hoomd-examples repository.
Placing barrier particles¶
You can create arbitrarily shaped barriers out of particles. This contrived example places particles on a sine wave in a 2D box of length L:
[4]:
L = 20
N_barrier = 100
x_barrier = numpy.linspace(start=-L / 2, stop=L / 2, endpoint=False, num=N_barrier)
Choose the period of the sine wave commensurate with the box length:
[5]:
periods = 2
barrier_amplitude = 4
x_scale = L / (periods * 2 * numpy.pi)
y_barrier = barrier_amplitude * numpy.sin(x_barrier / x_scale)
Place two barriers a given distance apart:
[6]:
separation = 2
y_barrier_top = y_barrier + separation + barrier_amplitude
y_barrier_bottom = y_barrier - separation - barrier_amplitude
position_barrier_top = numpy.stack(
(x_barrier, y_barrier_top, numpy.zeros(N_barrier)), axis=-1
)
position_barrier_bottom = numpy.stack(
(x_barrier, y_barrier_bottom, numpy.zeros(N_barrier)), axis=-1
)
box_height = barrier_amplitude * 4 + separation + 2
Placing mobile particles¶
You must place the mobile particles on the correct side of the barrier. This example places all particles on the line y=0:
[7]:
N_mobile = int(numpy.ceil(L / 1))
y_mobile = numpy.zeros(N_mobile)
x_mobile = (
numpy.linspace(start=-L / 2, stop=L / 2, endpoint=False, num=N_mobile) + 1 / 2
)
position_mobile = numpy.stack((x_mobile, y_mobile, numpy.zeros(N_mobile)), axis=-1)
Combine all particles in the simulation state¶
Make a GSD file containing all the particles:
[8]:
frame = gsd.hoomd.Frame()
frame.particles.N = N_mobile + N_barrier * 2
frame.particles.position = numpy.concatenate(
(position_mobile, position_barrier_top, position_barrier_bottom)
)
Give the mobile particles and the barrier particles separate types:
[9]:
frame.particles.types = ['mobile', 'barrier']
frame.particles.typeid = numpy.concatenate(
(numpy.tile(0, N_mobile), numpy.tile(1, N_barrier), numpy.tile(1, N_barrier)),
)
Set the box parameters:
[10]:
frame.configuration.box = [L, box_height, 0, 0, 0, 0]
[11]:
with gsd.hoomd.open(name='initial_state.gsd', mode='x') as f:
f.append(frame)
Perform HPMC simulations with barrier particles¶
Prepare a simulation of hard disks:
[12]:
simulation = hoomd.Simulation(device=hoomd.device.CPU(), seed=1)
simulation.create_state_from_gsd(filename='initial_state.gsd')
sphere = hoomd.hpmc.integrate.Sphere()
sphere.shape['mobile'] = dict(diameter=1.0)
sphere.shape['barrier'] = dict(diameter=1.0)
simulation.operations.integrator = sphere
Set the trial move size to 0 so the barrier does not move:
[13]:
sphere.d['barrier'] = 0
Run the simulation:
[14]:
simulation.run(10_000)
[15]:
render(simulation.state.get_snapshot())
[15]:
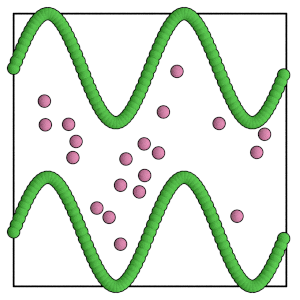
Carefully choose the move size for the mobile particles. Set the move size too large and a single trial move might pass completely to the other side of the barrier!
Perform MD simulations with barrier particles¶
In MD simulations, choose a pair potential interaction with a hard core to prevent the mobile particles from moving through the barrier:
[16]:
lj = hoomd.md.pair.LJ(nlist=hoomd.md.nlist.Cell(buffer=0.4))
lj.params[('mobile', 'mobile')] = dict(epsilon=1.0, sigma=1.0)
lj.r_cut[('mobile', 'mobile')] = 2.5
lj.params[('mobile', 'barrier')] = dict(epsilon=1.0, sigma=1.0)
lj.r_cut[('mobile', 'barrier')] = 2 ** (1 / 6)
Disable barrier-barrier particle interactions:
[17]:
lj.params[('barrier', 'barrier')] = dict(epsilon=0.0, sigma=0.0)
lj.r_cut[('barrier', 'barrier')] = 0
Integrate the equations of motion of the mobile particles:
[18]:
langevin = hoomd.md.methods.Langevin(filter=hoomd.filter.Type(types=['mobile']), kT=1.0)
Perform the MD simulation:
[19]:
simulation.operations.integrator = hoomd.md.Integrator(
dt=0.001, forces=[lj], methods=[langevin]
)
[20]:
simulation.run(10_000)
[21]:
render(simulation.state.get_snapshot())
[21]:
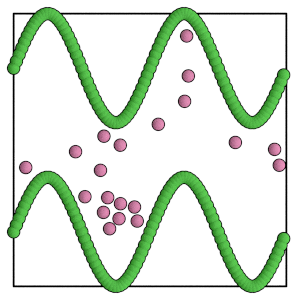
Scaling the box¶
When the barrier is made of particles, operations such as BoxResize and ConstantPressure can scale the barrier particles along with the mobile particles. Choose the appropriate values for filter and rescale_all to achieve this behavior.
[22]:
ramp = hoomd.variant.Ramp(A=0, B=1, t_start=simulation.timestep, t_ramp=20_000)
initial_box = simulation.state.box
final_box = hoomd.Box.from_box(initial_box)
final_box.volume = initial_box.volume / 2
box_resize = hoomd.update.BoxResize(
box1=initial_box,
box2=final_box,
variant=ramp,
trigger=hoomd.trigger.Periodic(1),
filter=hoomd.filter.All(),
)
simulation.operations.updaters.append(box_resize)
[23]:
simulation.run(20_000)
[24]:
render(simulation.state.get_snapshot())
[24]:
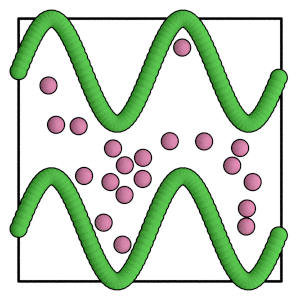
Conclusion¶
This section demonstrated how to use particles to create barriers in the simulation box for both HPMC and MD simulations. The next section explains wall geometries, another method to describe barriers.